GenomEast
PLATFORM LEADER
With over 20 years of experience, we offer a wide range of services to explore genomes, their expression and regulation, from quality control of starting samples to data analysis. These services are intended for the entire scientific community, national and international, public and private.
The platform is a member of the central core of the France Genomics National Research Infrastructure
The platform is IBISA certified.
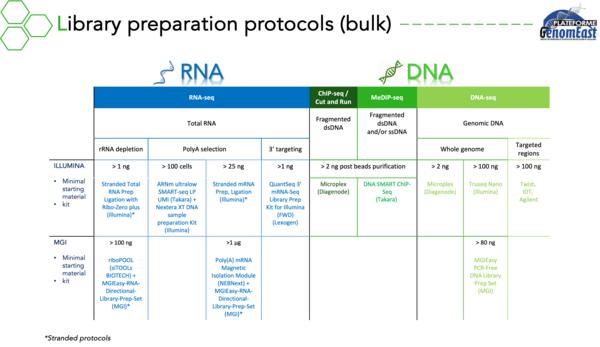
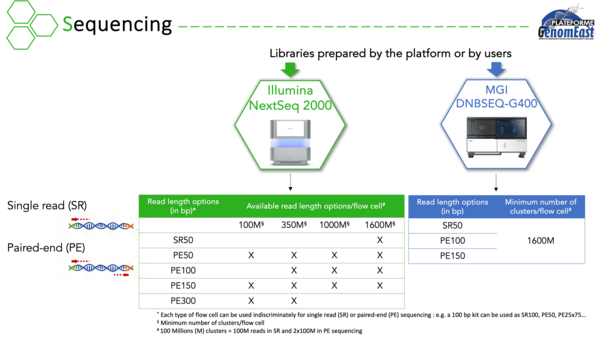
Currently equipped with an Illumina NextSeq 2000 and a MGI DNBSEQ-G400 sequencers, we are carrying out projects to analyse :
of the transcriptome :
- RNA-seq
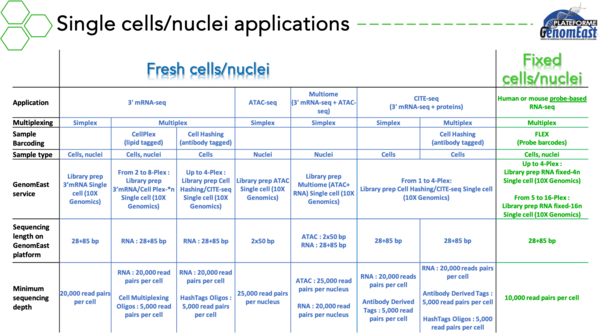
- Single cell RNA-seq (scRNA-seq)
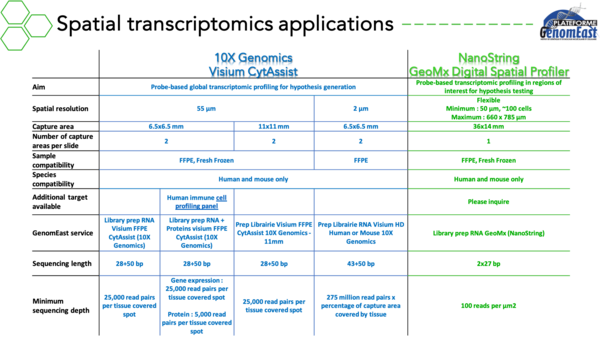
- Spatial transcriptomics
of the epigenome :
- ChIP-seq
- Cut & Run
- MeDIP-seq
- We also have experience in sequencing your ATAC-seq or Cut & Tag libraries
of the genome :
- Targeted regions of the genome (exome, regions of interest)
- Full genome sequencing
multiomics :
- Single Cell Multiome ATAC + Gene Expression
For these projects, we prepare and sequence your libraries, performing quality controls at each step. We can also perform the bioinformatics analysis of your data. We have expertise in a wide variety of applications and have developed dedicated bioinformatics pipelines to analyze the corresponding results.
Alternatively, you can use our sequencing capabilityto sequence your own libraries as long as they are compatible with Illumina technology.
How to submit a project to GenomEast?
Via our web interface
All our services are governed by the general terms and conditions of sale, which take precedence over any conditions of purchase, unless otherwise agreed in writing by us. You can also consult our Data Management Plan.
We regularly share our knowledge and expertise through various courses and training sessions, including:
- The "High Throughput Approaches" course at the Ecole Supérieure de Biotechnologie de Strasbourg (ESBS)
- The "Omics 2" course of the Master 2 Biology-Health, Biomedicine Research course at the University of Strasbourg
- The PhD Program of the IMCBio University Research School in Strasbourg
- The DU "High-throughput sequencing and rare diseases" at the University of Dijon
- The AVIESAN IFB Inserm bioinformatics school
- The organisation of training courses in bioinformatics for high-throughput sequencing with Inserm and the CNRS
Other equipments:
- High-throughput sequencing: iSeq100 (Illumina)
- Spatial transcriptomics: GeoMx Digital Spatial Profiler (NanoString)
- Quality control : Varioskan Flash Reader and Qubit Fluorometer (Thermo Fisher), 2100 Bioanalyzer and AATI Fragment Analyzer (Agilent)
- Sonicator : E220 AFA system (Covaris)
- IT Infrastructure : computing power (Dell): 592 cores, storage (Lenovo, GPFS): 750 TB
France Génomique
Our platform has been a partner of the France Genomics network since the initial selection of the project within the framework of the "Investissements d'avenir" in 2011. This network brings together and shares the resources of the main French genomics and bioinformatics platforms.
Fondation maladies rares
Since 2011, we have been one of the partner platforms of the Fondation maladies rares for the implementation of the "GenOmics: high throughput sequencing & rare diseases" programme, which aims to unravel the genetic and molecular basis of rare diseases using high throughput sequencing.
IBiSA and CoRTecS
Since 2008, we have been part of the national network of platforms and resource centers in biology, health, and agronomy of the GIS IBiSA, and since 2021, part of the regional CoRTecS network, which brings together the scientific research and service platforms of the University of Strasbourg.
New applications and technologies
Over the past years, we have gained a strong expertise in single-cell sequencing technologies (Research Expertise 2021 prize from the University of Strasbourg). We further implement new applications each year. At the same time, we are implementing two complementary technologies dedicated to spatial transcriptomics: Visium (10X Genomics) and GeoMx (NanoString).
Publications
2025
Pre-publication, Working Document
EXPLICIT CORRECTION OF SEVERELY NON-UNIFORM DISTRIBUTIONS OF CRYO-EM VIEWS
- Charles Barchet
- Ottilie von Loeffelholz
- Roberto Bahena-Ceron
- Bruno Klaholz
- Alexandre Urzhumtsev
2024
Article in a journal
A Gain-of-Function Mutation in the Ca2+ Channel ORAI1 Causes Stormorken Syndrome with Tubular Aggregates in Mice
- Laura Pérez-Guàrdia
- Emma Lafabrie
- Nadège Diedhiou
- Coralie Spiegelhalter
- Jocelyn Laporte
- Johann Böhm
Cells ; Volume: 13 ; Page: 1829
Pre-publication, Working Document
Classification of human actin pathological variants using C. elegans CRISPR-generated models
- Théo Hecquet
- Nadine Arbogast
- Delphine Suhner
- Anaïs Goetz
- Grégory Amann
- Selin Yürekli
- Fiona Marangoni
- Johannes N Greve
- Nataliya Di Donato
- Anne-Cécile Reymann
Article in a journal
Exome sequencing in undiagnosed congenital myopathy reveals new genes and refines genes–phenotypes correlations
- Yvan de Feraudy
- Marie Vandroux
- Norma Beatriz Romero
- Raphaël Schneider
- Safaa Saker
- Anne Boland
- Jean-François Deleuze
- Jean‐françois Deleuze
- Valérie Biancalana
- Johann Böhm
- Jocelyn Laporte
Genome Medicine ; Volume: 16 ; Page: 87
2022
Article in a journal
SAGA-Dependent Histone H2Bub1 Deubiquitination Is Essential for Cellular Ubiquitin Balance during Embryonic Development
- Farrah El-Saafin
- Didier Devys
- Steven Johnsen
- Stéphane Vincent
- László Tora
International Journal of Molecular Sciences ; Volume: 23 ; Page: 7459
2021
Chapter of the book
Practical Aspects of Super-Resolution Imaging and Segmentation of Macromolecular Complexes by dSTORM
- Leonid Andronov
- Jean-Luc Vonesch
- Bruno Klaholz
Multiprotein Complexes ; Volume: 2247 ; Page: 271-286
Pre-publication, Working Document
DetecDiv, a deep-learning platform for automated cell division tracking and replicative lifespan analysis
- Théo Aspert
- Didier Hentsch
- Gilles Charvin
2020
Article in a journal
Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders
- Manon Boivin
- Véronique Pfister
- Angeline Gaucherot
- Frank Ruffenach
- Luc Negroni
- Chantal Sellier
- Nicolas Charlet‐berguerand
EMBO Journal ; Volume: 39 ; Page: e100574
2019
Article in a journal
A cell cycle-coordinated Polymerase II transcription compartment encompasses gene expression before global genome activation
- Yavor Hadzhiev
- Haseeb Qureshi
- Lucy Wheatley
- Ledean Cooper
- Aleksandra Jasiulewicz
- Huy van Nguyen
- Joseph Wragg
- Divyasree Poovathumkadavil
- Sascha Conic
- Sarah Bajan
- Attila Sik
- György Hutvàgner
- Laszlo Tora
- Agnieszka Gambus
- John Fossey
- Ferenc Müller
Nature Communications ; Volume: 10
Pre-publication, Working Document
MEIOSIS INITIATES IN THE FETAL OVARY OF MICE LACKING ALL RETINOIC ACID RECEPTOR ISOTYPES
- Nadège Vernet
- Manuel Mark
- Diana Condrea
- Betty Féret
- Muriel Klopfenstein
- Violaine Alunni
- Marius Teletin
- Norbert B Ghyselinck